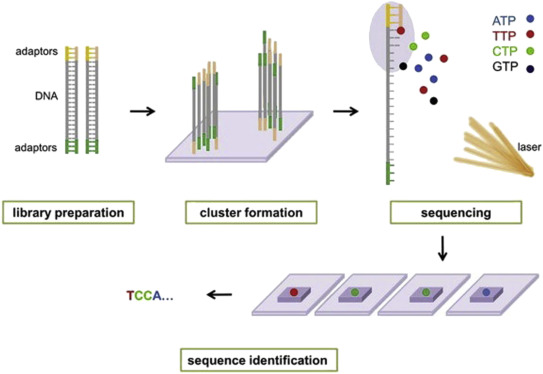
paired end sequencing wikipedia
Combining data from mate pair sequencing with those from short-insert paired-end reads provides increased information for maximising sequencing coverage across a genome 1. Paired end runs give additional positioning information in the genome making it a good choice for de novo genome assembly as well as making it easier to resolve structural re-arrangements.

What Is Mate Pair Sequencing For
Learn about the difference between Paired-End and Single-Run sequencing and why the former creates more precise alignments than the latter especiall.

. Mate pair sequencing is used for various applications applications including. This can be very helpful e. Typical experimental design advice for expression analyses using RNA-seq generally assumes that single-end reads provide robust gene-level expression estimates in a cost-effective manner and that the additional benefits obtained from paired-end sequencing are not worth the additional cost.
Mate pair sequencing is used for various applications applications including. For sequencing projects that require higher accuracy such as studies of alternate splicing 40 million to 60 million paired-end reads will provide better results. Combining data generated from mate pair library sequencing with that from short-insert paired-end reads provides a powerful combination of read lengths for maximal sequencing coverage across the genome.
As hmon stated most programs will have parameters to deal with paired-end sequencing and. For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. For mRNA-Seq library prep use.
Configure the system to sequence a trio in one day or up to 48 genomes in 2 days for the most comprehensive coverage. Mate pair sequencing involves generating long-insert paired-end DNA libraries useful for a number of sequencing applications including. For more detailed analyses to determine for example allele-specific expression or expression of low-abundant transcripts 60 million to 100 million reads may be required.
Furthermore sequencing can involve single-end SE or paired-end PE readsPaired-end sequencing means sequencing both ends of the cDNA fragments and aligning the forward and. For your De novo genome assembly Fig. Now lets get started.
The reads were then mapped back to the reference using BWA aln and sampe. Type of Run Single Read SR or Paired End PE With single read runs the sequencing instrument reads from one end of a fragment to the other end. This allows more accurate mapping particularly of repetitive regions.
For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. For mRNA-Seq library prep use. Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms.
Ad Access more DNA discoveries than has ever before been possible with Sequencing. Just a simple mouth swab. It is capable of automated paired-end reads and up to 15 Gb per run delivering over 600 bases of sequence data per read.
Paired-end RNA sequencing RNA-Seq enables discovery applications such as detecting gene fusions in cancer and characterizing novel splice isoforms. The library prep kits that it uses are optimized for a variety of applications including targeted gene small genome and amplicon. The inner mate distance between the two reads is 200 bp creating an insert size of 400 bp.
Another consideration is whether to generate a strand-specific library that retains the orientation of the original RNA transcript which may be critical to identify antisense or non-coding RNA. For paired-end RNA-Seq use the following kits with an alternate fragmentation protocol followed by standard Illumina paired-end cluster generation and sequencing. However in many cases eg with Illumina NextSeq and NovaSeq.
The grey represents the fragment and each end of the fragment is sequenced. The MiSeq System facilitates your research with a wide range of sequencing applications. Theres also a great animation here that illustrates the concept of Illumina paired end sequencing.

Hi C Genomic Analysis Technique Wikipedia
How Has Sequencing Technology Advanced Since The Development Of The Process By Fred Sanger Quora
Difference Between Whole Genome Sequencing And Next Generation Sequencing Difference Between
Manipulating Ngs Data With Galaxy Galaxy Community Hub

Dna Sequencing Wikipedia Pdf Dna Sequencing Dna

Hi C Genomic Analysis Technique Wikipedia
Proximal End Bias From In Vitro Reconstituted Nucleosomes And The Result On Downstream Data Analysis Plos One

End Sequence Profiling Wikiwand

What Is Mate Pair Sequencing For
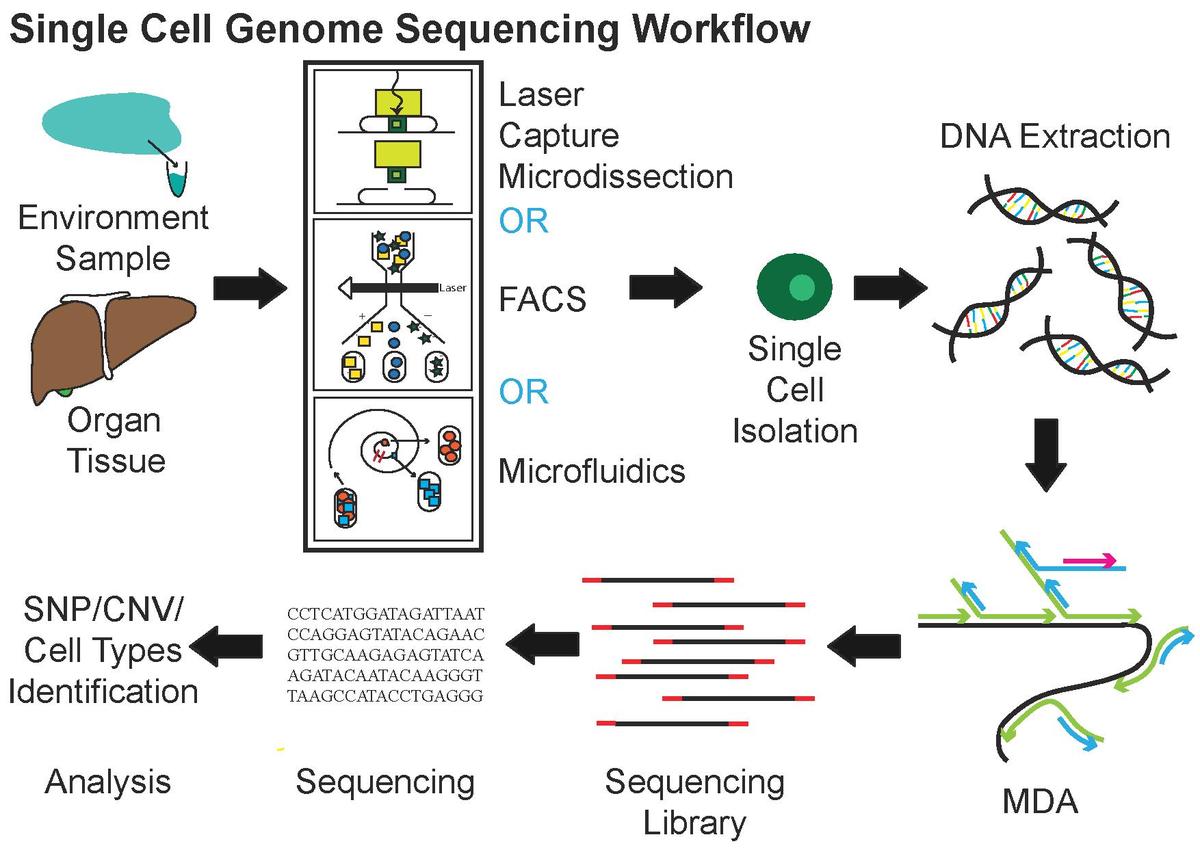
Chapter 1 Introduction To Single Cell Technology Fundamentals Of Scrnaseq Analysis